Innovative Chemical Engineering Simulations Unveil Breakthroughs in Pain Therapy Research

The building blocks of life — cells, biomolecules, proteins — have been viewed through microscopes since the 17th century. In the Department of Chemical Engineering at the University of California, Davis, the Ahn Lab, led by Assistant Professor Surl-Hee (Shirley) Ahn, is pioneering cutting-edge simulation tools that are changing how scientists can look at these molecules and the ways they move, grow and change.
Ahn uses molecular dynamics, or MD, simulations to view proteins, other biomolecules and materials at the atomic level as they are changing from one state to another, instead of observing the molecules in one state and then later after it has changed. MD simulations are more like making a movie rather than taking photos.
"The easiest way to think about it is we're trying to work with a computational microscope," Ahn said. "You can see protein structures experimentally, but you can only get snapshots, and you get static information. But with the aid of computer simulations, you can see how the structure moves with atomic resolution, and you get the dynamic information, which is not possible with experimental techniques."
The Right Tool for the Job
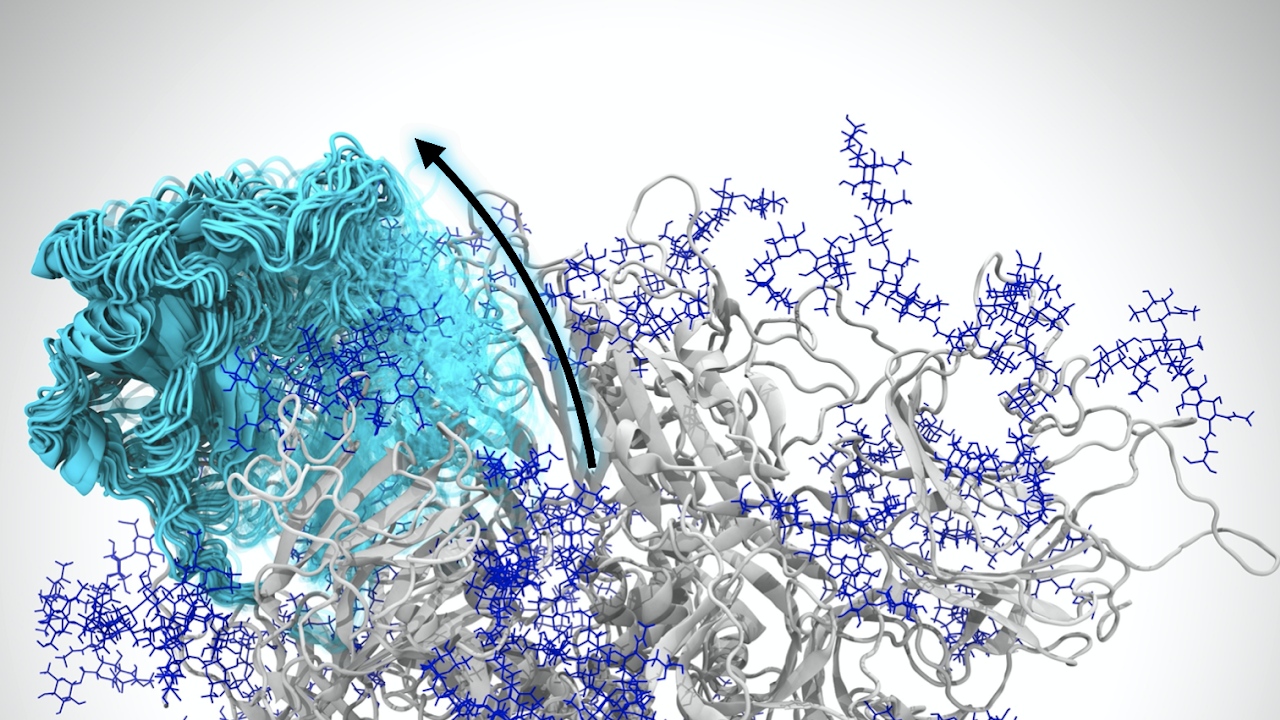
MD simulations were most recently highlighted in a research collaboration with the UC Davis School of Medicine's Department of Physiology and Membrane Biology. The research was published in the Journal of General Physiology on Feb. 5, revealing how a toxin derived from tarantulas binds to specific pain receptors in nerve cells, providing a blueprint for making new and better pain therapies.
The researchers sought to investigate protoxin-II, a peptide that comes from tarantula venom. Defined as smaller versions of proteins, peptides can act as transports or delivery systems for toxins. In this way, protoxin-II acts as a delivery system for the venom, targeting particular cells to get the desired effect, like paralysis.
Specifically, the researchers wanted to know how protoxin-II interacts with certain human molecules that generate electrical signals in cells called sodium-ion channels, which are involved in several physiological processes, including pain signaling.
The MD simulations, for which Ahn's lab provided the tools and analysis, revealed which residues and lipid contacts are important in the binding of protoxin-II and the sodium ion channel.
Although effective as a potential pain remedy, protoxin-II has shown adverse effects on rats and cannot be used in its current state as a pain therapy. These findings, however, could lead to researchers building peptides with similar properties to protoxin-II for pain therapies that would better target specific sodium-ion channels, making them more effective and with fewer side effects than other therapies like anesthesia and opioids. (The project is related to an NIH-funded initiative to find alternatives to opioids, for which UC Davis is contributing research.)
Crunching Time
Ahn's expertise in the weighted ensemble method, which she and her lab are continuously developing and improving, was also crucial in the research.
The method acts as a bridge between the timescales of MD simulations, which run using femtosecond (one-millionth of one-billionth of a second) time steps, and the timescales of biophysical processes that researchers are interested in, like seconds or hours. The process allows researchers to efficiently sample kinetic pathways, like a protein transitioning from an open to a closed state.
Igor Vorobyov, an assistant professor of physiology and membrane biology and one of the project's lead researchers, says having Ahn's tools and expertise in this research was a huge advantage.
"For many processes we are interested in, we cannot use conventional simulation methods because we are limited to a much shorter timescale," he said. "By combining the molecular dynamics simulations and the weighted ensemble method that Shirley pioneered, we can accelerate sampling of different protein structures, how they evolve in time, and how they interact with each other. That's a great advantage."
Current and Future Applications
Vorobyov notes that this is just the beginning of their collaboration and that with these tools and the knowledge of Ahn Lab, they can take their research to the next level.

"To be able to calculate the different energetics of interactions or why different proteins like or dislike each other, or how fast these peptides can come in and out are all very crucial for a better understanding of this process and for connecting simulations to larger scale models, which allow us to see not only the protein but the whole nerve cell and even the whole human body."
Ahn is applying MD simulations and the weighted ensemble methods to other areas, specifically diseases.
One of Ahn's lab members, Celeste Wan, who is earning a Ph.D. in chemical engineering, is investigating the relationship between alpha-synuclein, a protein implicated in Parkinson's disease, with small molecule inhibitors to find which residues are important in the binding and inhibition of the molecular level. Similar to the protoxin-II research, the project is in collaboration with the UC Davis School of Medicine's John Voss, a professor at the Department of Biochemistry and Molecular Medicine, and Qizhi Gong, a professor in the Department of Cell Biology and Human Anatomy.
Vorobyov sees unlimited potential for how Ahn's tools and methods can be applied and how they can impact research.
"This approach can be used for multiple kinds of systems, and hopefully, we can also provide something equal to what people do and see in experiments," he said. "With Shirley's approach, we should be able to do that."
